Summary of DNA methylation analysis
A brief summary of our DNA methylation analysis.
RRBS
We used RRBS (reduced representation bisulfite sequencing) to analyse the DNA methylation profiles affected by our experimental feed with different micronutrient compositions. RRBS utilises restriction enzymes to target CpG rich regions in the genome. We used two different enzymes for our RRBS analysis.
Restriction enzymes used for RRBS
- MspI (recognition sequence :
5' CCGG) - TaqI (recognition sequence :
5' TCGA)
RRBS samples
We collected 21 liver samples at the final harvest stage for RRBS sequencing.
The number of samples for each group
- L1 diet: 9 samples
- L2 diet: 6 samples
- L3 diet: 6 samples
| No | Name | Diet | Sex |
|---|---|---|---|
| 1 | L1 | L2 | M |
| 2 | L6 | L3 | M |
| 3 | L7 | L3 | F |
| 4 | L11 | L1 | M |
| 5 | L12 | L1 | F |
| 6 | L13 | L1 | F |
| 7 | L14 | L1 | F |
| 8 | L16 | L2 | F |
| 9 | L18 | L2 | M |
| 10 | L20 | L2 | F |
| 11 | L22 | L1 | M |
| 12 | L23 | L1 | M |
| 13 | L24 | L1 | M |
| 14 | L26 | L3 | M |
| 15 | L28 | L3 | F |
| 16 | L30 | L3 | F |
| 17 | L32 | L2 | M |
| 18 | L33 | L2 | F |
| 19 | L38 | L3 | F |
| 20 | L42 | L1 | F |
| 21 | L43 | L1 | F |
Definition of genomic regions
Functions of DNA methylation can be different depending on the types of regions where methylation occurs. We split the genome into seven different regions for our RRBS analysis.
Genomic regions for RRBS read mapping
- Exon
- Intron
- P250 (proximal promoter)
- P1K (promoter)
- P6K (distal promoter)
- Flanks (potential enhancer region)
- IGR (intergenic region)

Bioinformatics pipelne for RRBS
We used various bioinformatics algorithms and methods to analyse our RRBS samples. The following tools were those we used in our main RRBS pipeline.
- FastQC (quality control)
- Trim Galore! (adapter trimming)
- MultiQC (quality control report)
- Bismark (read alignment)
- methylKit (differential methylation analysis)
Results
Overall diet effect
Unlike the RNA-seq samples, t-SNE clustering analysis showed no obvious separations of RRBS samples by diet.

Differentially methylated CpG sites
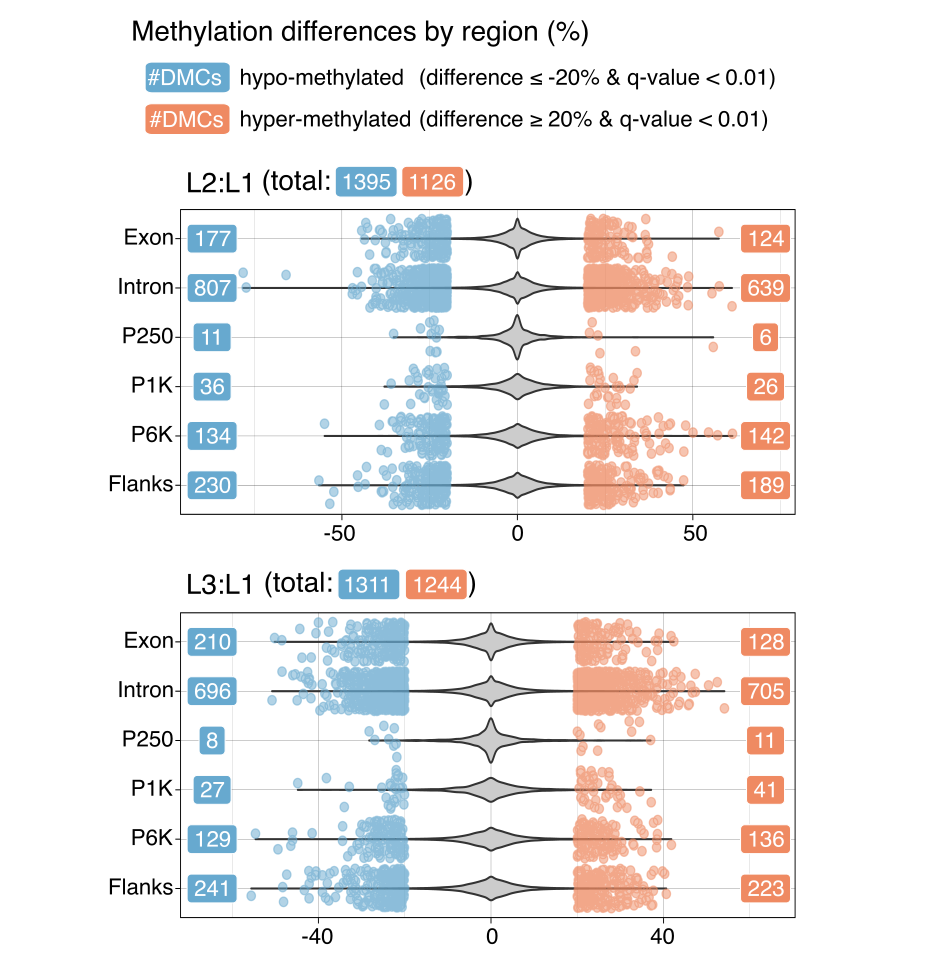
There were no noticeable differences between L2:L1 and L3:L1 as well as hypo- and hyper-methylation in terms of the number of DMCs.
Identified DMCs for L2:L1 and L3:L1
- L2 vs. L1: 2521 DMCs
- L3 vs. L1: 2555 DMCs
See What are DMCs? for more details about DMCs.
Significantly affected biological pathways
ORA (over representation analysis) on KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways showed that micronutrient supplement significantly affected DNA methylation profiles in cell-adhesion and cell-signalling pathways.
| Dataset | Pathway | KEGG ID | ORA (gene ratio) |
|---|---|---|---|
| L2:L1 | Cell adhesion molecules (CAMs) | sasa04514 | RS+GB (38/616), P+GB (32/517), Gene body (31/420), Intron (26/339) |
| Glycosaminoglycan degradation | sasa00531 | P1K (2/21) | |
| Mitophagy - animal | sasa04137 | Promoter (7/99), P1K (3/21) | |
| Apelin signalling pathway | sasa04371 | P+GB (28/517) | |
| Starch and sucrose metabolism | sasa00500 | P6K (4/74) | |
| L3:L1 | Cell adhesion molecules (CAMs) | sasa04514 | RS+GB (37/573), P+GB (30/465), Gene body (27/388), Intron (27/320) |
| ECM-receptor interaction | sasa04512 | Intron (15/320) |
Page links
Overview
DMC: differentially methylated CpG site
DMG: differentially methylated gene
Leave a comment